Effects of climate change on population interaction: Coupled integro-differential equations
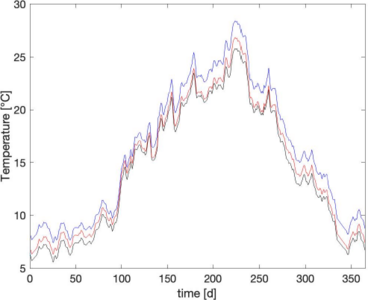
Stability of food webs under environmental changes like e.g. climate change depends on the effect of a time-dependent parameter like temperature on population level and community level. As predator-prey relationships depend on individual size, all populations involved must be size-structured when it comes to quantification. In the framework of a COMSOL model, the range of possible individual sizes is represented as the geometry, and a transport and reaction equation is implemented. Here, reproduction is a source, growth is transport and mortality is a sink. Predatory success shapes the predator’s growth and the prey’s mortality. The theoretical background was explored years ago for a single cannibalistic population (Claessen and De Roos, 2003) or a predatory population with an unstructured prey (van de Wolfshaar et al., 2006). However, quantifying size-specific predation (including cannibalism) for two structured populations results in coupled integrals over different geometries. Here, COMSOL allows for a new synthesis of monitoring data and field experiments, an analysis of all types of interactions over time and projection of climate change scenarios to identify vulnerability. The analysis presented considers the interaction of Perca fluviatilis, Neogobius melanostomus, and Dikerogammarus villosus in river Rhine. It is based on population specific analysis (Thapa, M. S., 2021; Minor R., 2020) and interaction analysis (Lazarev, K., 2021). Interaction analysis indicated that in near future P. fluviatilis will probably satisfy its demands where D. villosus might be partially replaced by N. melanostomus. Additionally, the frequency and intensity of extreme weather conditions will be decisive in presence and absence of specific species. A fully coupled food web model combined with monitoring data should further explore these findings.
下载
- moenickes_10501_poster.pdf - 0.74MB
